mebioda
High-throughput DNA sequencing
Data centric view of genome resequencing workflow
- (semi-)raw reads come out of lab as FASTQ
- reads are assembled to a FASTA file
- reads are aligned to a (pseudo-)reference as SAM/BAM/CRAM
- alignment is viewed in genome browser, with additional tracks as BED
- variants are called (e.g. as VCF/BCF), consensus is computed (e.g. as FASTA)
- consensus is annotated as GFF
The FASTA format
- The simplest, and therefore most common sequence format
- Holds DNA/RNA/AA sequence data in IUPAC single characters
- Flat text, can be read by most bioinformatics toolkits
- Seen in Genomic Architectures, used for online BLAST
- Syntax:
>Definition_line agctcagactacgactacgcatcagcga
Extensions:
- The first words in the definition line are often used to embed data, e.g. identifiers
- Sequence quality is sometimes written to separate FASTA-like files with records in the same order (and same length)
- Can be one or multiple records, which might be aligned, in which case there
might be gap
-characters - Lower case and capitalized are sometimes used to embed information such as base quality or strandedness
The FASTQ format
- The initial mountain of data to deal with
- Sequential format returned by most HTS platforms
- Flat text, can be read by most bioinformatics toolkits
- Includes base calling quality scores
Record layout:
@+identifier - note paired-end sequencing- Sequence data - IUPAC single character nucleotides
+- separator between sequence and quality- Quality lines - map phred scores to ASCII characters
Example:
@FAKE0005
ACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACG
+
@ABCDEFGHIJKLMNOPQRSTUVWXYZ[\]^_`abcdefghijklmnopqrstuvwxyz{|}~
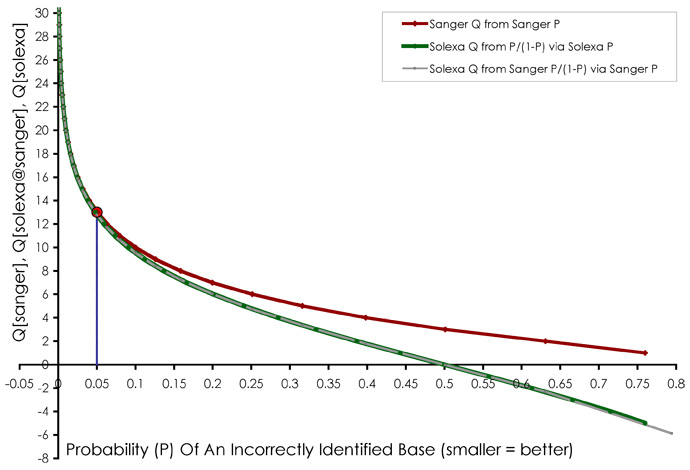
Quality (phred) scores
Phred quality scores Q are defined as a property which is logarithmically related to the base-calling error probabilities P.
Q = -10 log10 P
For example, if Phred assigns a quality score of 30 to a base, the chances that this base is called incorrectly are 1 in 1000.
| Phred Quality Score | Probability of incorrect base call | Base call accuracy |
|---|---|---|
| 10 | 1 in 10 | 90% |
| 20 | 1 in 100 | 99% |
| 30 | 1 in 1000 | 99.9% |
| 40 | 1 in 10,000 | 99.99% |
| 50 | 1 in 100,000 | 99.999% |
| 60 | 1 in 1,000,000 | 99.9999% |
Phred score encoding
Different platforms map phred scores in different ways to ASCII:
- sanger: 33..126
- solexa: 59..126
- illumina: 64..126
| code | char | code | char | code | char | code | char | code | char | code | char |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 33 | ! | 49 | 1 | 65 | A | 81 | Q | 97 | a | 113 | q |
| 34 | ” | 50 | 2 | 66 | B | 82 | R | 98 | b | 114 | r |
| 35 | # | 51 | 3 | 67 | C | 83 | S | 99 | c | 115 | s |
| 36 | $ | 52 | 4 | 68 | D | 84 | T | 100 | d | 116 | t |
| 37 | % | 53 | 5 | 69 | E | 85 | U | 101 | e | 117 | u |
| 38 | & | 54 | 6 | 70 | F | 86 | V | 102 | f | 118 | v |
| 39 | ’ | 55 | 7 | 71 | G | 87 | W | 103 | g | 119 | w |
| 40 | ( | 56 | 8 | 72 | H | 88 | X | 104 | h | 120 | x |
| 41 | ) | 57 | 9 | 73 | I | 89 | Y | 105 | i | 121 | y |
| 42 | * | 58 | : | 74 | J | 90 | Z | 106 | j | 122 | z |
| 43 | + | 59 | ; | 75 | K | 91 | [ | 107 | k | 123 | { |
| 44 | , | 60 | < | 76 | L | 92 | \ | 108 | l | 124 | | |
| 45 | - | 61 | = | 77 | M | 93 | ] | 109 | m | 125 | } |
| 46 | . | 62 | > | 78 | N | 94 | ^ | 110 | n | 126 | ~ |
| 47 | / | 63 | ? | 79 | O | 95 | _ | 111 | o | ||
| 48 | 0 | 64 | @ | 80 | P | 96 | ` | 112 | p |
The SAM/BAM/CRAM format
- Format to represent (FASTQ) reads aligned to a reference sequence
- Textual (SAM) and binary representations (BAM)
- Binary representation further compressed as CRAM
- Created using reference mapping tools such as BWA
- Accessed using tools such as samtools, picard, and scripting toolkits built on top of these (e.g. PySAM)
Example alignment:
Coor 12345678901234 5678901234567890123456789012345
ref AGCATGTTAGATAA**GATAGCTGTGCTAGTAGGCAGTCAGCGCCAT
+r001/1 TTAGATAAAGGATA*CTG
+r002 aaaAGATAA*GGATA
+r003 gcctaAGCTAA
+r004 ATAGCT..............TCAGC
-r003 ttagctTAGGC
-r001/2 CAGCGGCAT
- Read
r001/1andr001/2constitute a read pair r003is a chimeric readr004represents a split alignment
SAM representation
Example:
@HD VN:1.5 SO:coordinate
@SQ SN:ref LN:45
r001 99 ref 7 30 8M2I4M1D3M = 37 39 TTAGATAAAGGATACTG *
r002 0 ref 9 30 3S6M1P1I4M * 0 0 AAAAGATAAGGATA *
r003 0 ref 9 30 5S6M * 0 0 GCCTAAGCTAA *
r004 0 ref 16 30 6M14N5M * 0 0 ATAGCTTCAGC *
r003 2064 ref 29 17 6H5M * 0 0 TAGGC *
r001 147 ref 37 30 9M = 7 -39 CAGCGGCAT *
Mandatory columns:
| No. | Name | Description |
|---|---|---|
| 1 | QNAME | Query NAME of the read or the read pair |
| 2 | FLAG | Bitwise FLAG (pairing, strand, mate strand, etc.) |
| 3 | RNAME | Reference sequence NAME |
| 4 | POS | 1-Based leftmost POSition of clipped alignment |
| 5 | MAPQ | MAPping Quality (Phred-scaled) |
| 6 | CIGAR | Extended CIGAR string (operations: MIDNSHP) |
| 7 | MRNM | Mate Reference NaMe (‘=’ if same as RNAME) |
| 8 | MPOS | 1-Based leftmost Mate POSition |
| 9 | ISIZE | Inferred Insert SIZE |
| 10 | SEQ | Query SEQuence on the same strand as the reference |
| 11 | QUAL | Query QUALity (ASCII-33=Phred base quality) |
What is a CIGAR?
The sequence being aligned to a reference may have additional bases that are not in the reference or may be missing bases that are in the reference. The CIGAR string is a sequence of base lengths and the associated operation. They are used to indicate things like which bases align (either a match/mismatch) with the reference, are deleted from the reference, and are insertions that are not in the reference. For example:
RefPos: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
Reference: C C A T A C T G A A C T G A C T A A C
Read: ACTAGAATGGCT
Aligning these two:
RefPos: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
Reference: C C A T A C T G A A C T G A C T A A C
Read: A C T A G A A T G G C T
With the alignment above, you get:
POS: 5
CIGAR: 3M1I3M1D5M
The POS indicates that the read aligns starting at position 5 on the reference. The CIGAR says that the first 3 bases in the read sequence align with the reference. The next base in the read does not exist in the reference. Then 3 bases align with the reference. The next reference base does not exist in the read sequence, then 5 more bases align with the reference. Note that at position 14, the base in the read is different than the reference, but it still counts as an M since it aligns to that position.
Bitwise flags

- The Bits column shows all the 12 switches that can possibly be set
- SAM property shows the meaning of the one switch that is set in the corresponding Bits column
- The first column shows the number of combinations possible, counting all the switches from right to the current one, as 2 raised to the power of x, where x is the position of the switch
- The Value (Decimal) column shows what that number of combinations is for normal people
- The Value (Hex) column shows the same number if you were counting with 16 fingers
| 2x | Value (Hex) | Value (Decimal) | Bits | SAM property |
|---|---|---|---|---|
| 20 | 1 | 1 | 000000000001 | are there multiple fragments? |
| 21 | 2 | 2 | 000000000010 | are all fragments properly aligned? |
| 22 | 4 | 4 | 000000000100 | is this fragment unmapped? |
| 23 | 8 | 8 | 000000001000 | is the next fragment unmapped? |
| 24 | 10hex | 16 | 000000010000 | is this query the reverse strand? |
| 25 | 20hex | 32 | 000000100000 | is the next fragment the reverse strand? |
| 26 | 40hex | 64 | 000001000000 | is this the 1st fragment? |
| 27 | 80hex | 128 | 000010000000 | is this the last fragment? |
| 28 | 100hex | 256 | 000100000000 | is this a secondary alignment? |
| 29 | 200hex | 512 | 001000000000 | did this read fail quality controls? |
| 2A | 400hex | 1024 | 010000000000 | is this read a PCR or optical duplicate? |
| 2B | 800hex | 2048 | 100000000000 | supplementary alignment |
The values in the FLAG column (2) correspond to bitwise flags as follows:
| FLAG | hex | Explanation |
|---|---|---|
| 99 | 0x63 | first/next is reverse-complemented/properly aligned/multiple segments |
| 0 | no flags set, thus a mapped single segment | |
| 2064 | 0x810 | supplementary/reversecomplemented |
| 147 | 0x93 | last (second of a pair)/reverse-complemented/properly aligned/multiple segments |
Using the flags, we can filter the reads in a BAM file using samtools:
$ samtools view -f 4 file.bam > unmapped.sam
The BED format
If you load a SAM/BAM file in a genome browser (e.g. UCSC) you might want to load additional ‘tracks’ alongside the alignment. For this the BED format is used.
track name=pairedReads description="Clone Paired Reads" useScore=1
chr22 1000 5000 cloneA 960 + 1000 5000 0 2 567,488, 0,3512
chr22 2000 6000 cloneB 900 - 2000 6000 0 2 433,399, 0,3601
- The name of the chromosome (e.g. chr3, chrY, chr2_random) or scaffold (e.g. scaffold10671).
- The starting position of the feature in the chromosome or scaffold. The first base in a chromosome is numbered 0.
- The ending position of the feature in the chromosome or scaffold. The chromEnd base is not included in the display of the feature. For example, the first 100 bases of a chromosome are defined as chromStart=0, chromEnd=100, and span the bases numbered 0-99.
- Defines the name of the BED line. This label is displayed to the left of the BED line in the Genome Browser window when the track is open to full display mode or directly to the left of the item in pack mode.
- A score between 0 and 1000. If the track line useScore attribute is set to 1 for this annotation data set, the score value will determine the level of gray in which this feature is displayed (higher numbers = darker gray).
- Defines the strand. Either “.” (=no strand) or “+” or “-“.
- The starting position at which the feature is drawn thickly (for example, the start codon in gene displays). When there is no thick part, thickStart and thickEnd are usually set to the chromStart position.
- The ending position at which the feature is drawn thickly (for example the stop codon in gene displays).
- An RGB value of the form R,G,B (e.g. 255,0,0). If the track line itemRgb attribute is set to “On”, this RBG value will determine the display color of the data contained in this BED line. NOTE: It is recommended that a simple color scheme (eight colors or less) be used with this attribute to avoid overwhelming the color resources of the genome Browser and your Internet browser.
- The number of blocks (exons) in the BED line.
- A comma-separated list of the block sizes. The number of items in this list should correspond to blockCount.
- A comma-separated list of block starts. All of the blockStart positions should be calculated relative to chromStart. The number of items in this list should correspond to blockCount.
The VCF/BCF format
- Format for variants (SNPs, indels, microsats) computed from a SAM/BAM/CRAM file
- Concise, good for resequencing projects, but lossy
- Text and binary version
##fileformat=VCFv4.2
##fileDate=20090805
##source=myImputationProgramV3.1
##reference=file:///seq/references/1000GenomesPilot-NCBI36.fasta
##contig=<ID=20,length=62435964,assembly=B36,md5=f126cdf8a6e0c7f379d618ff66beb2da,species="Homo sapiens",taxonomy=x>
##phasing=partial
##INFO=<ID=NS,Number=1,Type=Integer,Description="Number of Samples With Data">
##INFO=<ID=DP,Number=1,Type=Integer,Description="Total Depth">
##INFO=<ID=AF,Number=A,Type=Float,Description="Allele Frequency">
##INFO=<ID=AA,Number=1,Type=String,Description="Ancestral Allele">
##INFO=<ID=DB,Number=0,Type=Flag,Description="dbSNP membership, build 129">
##INFO=<ID=H2,Number=0,Type=Flag,Description="HapMap2 membership">
##FILTER=<ID=q10,Description="Quality below 10">
##FILTER=<ID=s50,Description="Less than 50% of samples have data">
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
##FORMAT=<ID=GQ,Number=1,Type=Integer,Description="Genotype Quality">
##FORMAT=<ID=DP,Number=1,Type=Integer,Description="Read Depth">
##FORMAT=<ID=HQ,Number=2,Type=Integer,Description="Haplotype Quality">
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA00001 NA00002 NA00003
20 14370 rs6054257 G A 29 PASS NS=3;DP=14;AF=0.5;DB;H2 GT:GQ:DP:HQ 0|0:48:1:51,51 1|0:48:8:51,51 1/1:43:5:.,.
20 17330 . T A 3 q10 NS=3;DP=11;AF=0.017 GT:GQ:DP:HQ 0|0:49:3:58,50 0|1:3:5:65,3 0/0:41:3
20 1110696 rs6040355 A G,T 67 PASS NS=2;DP=10;AF=0.333,0.667;AA=T;DB GT:GQ:DP:HQ 1|2:21:6:23,27 2|1:2:0:18,2 2/2:35:4
20 1230237 . T . 47 PASS NS=3;DP=13;AA=T GT:GQ:DP:HQ 0|0:54:7:56,60 0|0:48:4:51,51 0/0:61:2
20 1234567 microsat1 GTC G,GTCT 50 PASS NS=3;DP=9;AA=G GT:GQ:DP 0/1:35:4 0/2:17:2 1/1:40:3
- a good simple SNP
- a possible SNP that has been filtered out because its quality is below 10
- a site at which two alternate alleles are called, with one of them (T) being ancestral (possibly a reference sequencing error)
- a site that is called monomorphic reference (i.e. with no alternate alleles)
- a microsatellite with two alternative alleles, one a deletion of 2 bases (TC), and the other an insertion of one base (T).
Genotype data are given for three samples, two of which are phased and the third unphased, with per sample genotype quality, depth and haplotype qualities (the latter only for the phased samples) given as well as the genotypes. The microsatellite calls are unphased.
The GFF format
Once a plausible sequence has been generated (e.g. by doing a de novo assembly or by computing a consensus from a genome alignment) the next step might be to annotate it, i.e. predict ORFs and scan databases for homologies. Such genome annotations are stored in GFF files:
contig_1 maker gene 1797 7025 . + . ID=CR513_016834;Name=CR513_016834;Alias=maker-contig_1-snap-gene-0.0;Note=Similar to SCAI: Protein SCAI (Homo sapiens);
contig_1 maker five_prime_UTR 1797 1954 . + . ID=CR513_016834-RA:five_prime_utr;Parent=CR513_016834-RA;
contig_1 maker CDS 1955 2095 . + 0 ID=CR513_016834-RA:cds;Parent=CR513_016834-RA;
contig_1 maker CDS 2223 2359 . + 0 ID=CR513_016834-RA:cds;Parent=CR513_016834-RA;
contig_1 maker CDS 6748 6979 . + 1 ID=CR513_016834-RA:cds;Parent=CR513_016834-RA;
contig_1 maker three_prime_UTR 6980 7025 . + . ID=CR513_016834-RA:three_prime_utr;Parent=CR513_016834-RA;
- name of the chromosome or scaffold; chromosome names can be given with or without the ‘chr’ prefix. Important note: the seqname must be one used within Ensembl, i.e. a standard chromosome name or an Ensembl identifier such as a scaffold ID, without any additional content such as species or assembly. See the example GFF output below.
- name of the program that generated this feature, or the data source (database or project name)
- feature type name, e.g. Gene, Variation, Similarity
- start position of the feature, with sequence numbering starting at 1.
- end position of the feature, with sequence numbering starting at 1.
- a floating point value.
- defined as + (forward) or - (reverse).
- one of ‘0’, ‘1’ or ‘2’. ‘0’ indicates that the first base of the feature is the first base of a codon, ‘1’ that the second base is the first base of a codon, and so on.
- semicolon-separated list of tag-value pairs, providing additional information about each feature.
Submission to NCBI
See: WGS2NCBI