mebioda
Large phylogenies in biodiversity
- Why care about large phylogenies outside systematics?
- Which projects synthesize large phylogenies for re-use?
- How to use such big trees? What tooling exists?
- (Exercise) what’s the subtree for our crops?
Why care about large phylogenies outside systematics?
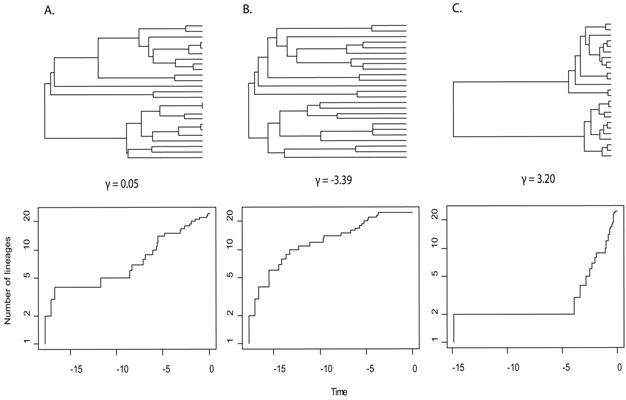
- The distribution of splits on a tree says something about diversification processes (example: LTT plot in context of GMYC species delimitation)
- The topology and branch lengths on a subtree say something about the phylogenetic diversity of the taxa in that subtree in comparison with other subtrees, as beta diversity (example: UniFrac index to compare sites pre- and post- DeepWater Horizon spill)
- Because taxa are non-independent from one another (all life is related), their characteristics cannot be analyzed as independent observations: we have to consider phylogeny as a source of covariation
Large, ad hoc phylogenies: Mammals
ORP Bininda-Emonds, et al. 2007. The delayed rise of present-day mammals. Nature 446: 507–512 doi:10.1038/nature05634

- Constructed by combining many, smaller input trees into a consensus-like (“supertree”) method
- Time-calibrated using molecules and fossils
- Reported result: mammals started diversifying earlier than thought
- Tree cited and re-used many times
Large, ad hoc phylogenies: Birds
W Jetz, et al. 2012. The global diversity of birds in space and time. Nature 491: 444–448 doi:10.1038/nature11631
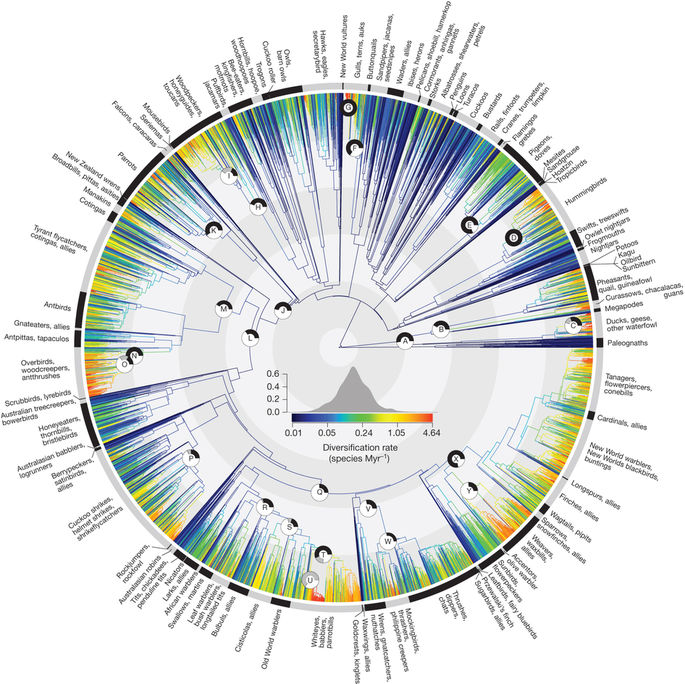
- Constructed mostly on the basis of molecular data
- Time-calibrated using relaxed molecular clocks
- Reported result: bird diversification rates increased 50MYA
- Tree cited and re-used many times, thanks to the accompanying data resource birdtree.org
Large, ad hoc phylogenies: Plants
AE Zanne, et al. 2014. Three keys to the radiation of angiosperms into freezing environments. Nature 506: 89–92 doi:10.1038/nature12872
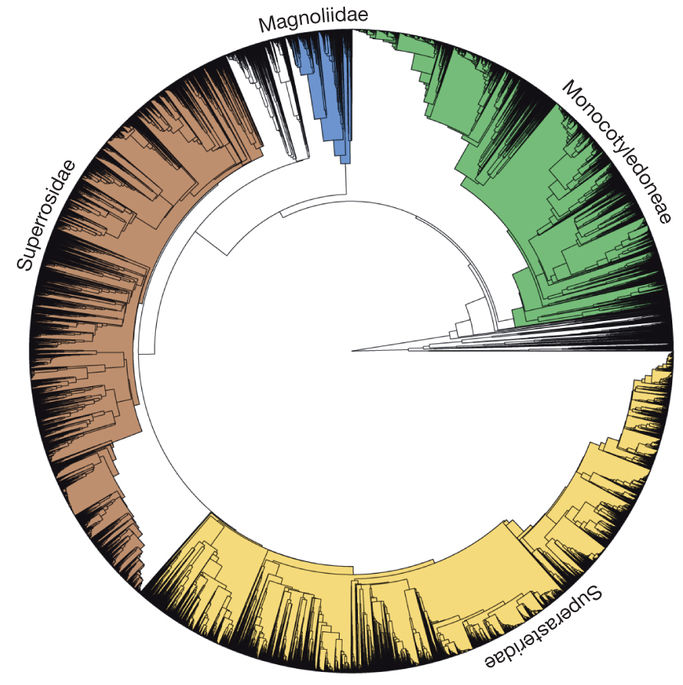
- Constructed using molecular data
- Combined with trait and ecological data
- Reported result: certain traits allowed plants to colonize freezing environments
Large, ongoing, phylogenetic projects
Apart from these ad hoc projects where a tree was published once, there are ongoing initiatives to periodically release estimates of phylogeny for a given taxonomic group and/or marker.
Examples from molecular biodiversity:
- SILVA - curated datasets of aligned small (16S/18S, SSU) and large subunit (23S/28S, LSU) ribosomal RNA (rRNA) sequences for all three domains of life (Bacteria, Archaea and Eukarya).
- GreenGenes - curated database of small (16S) subunit ribosomal near-full length sequences from the kingdoms Bacteria and Archaea.
Examples of species tree initiatives:
- Tree of Life Web project - systematics resource where clades are maintained by taxonomic experts. Hence, topology is based on the judgment of expert individuals.
- Angiosperm Phylogeny working Group (APG) - community effort to release classifications (now at version IV) of flowering plants on the basis of phylogenetic systematics.
- Open Tree of Life - open source project to develop an infrastructure for building supertree-like phylogenetic estimates.
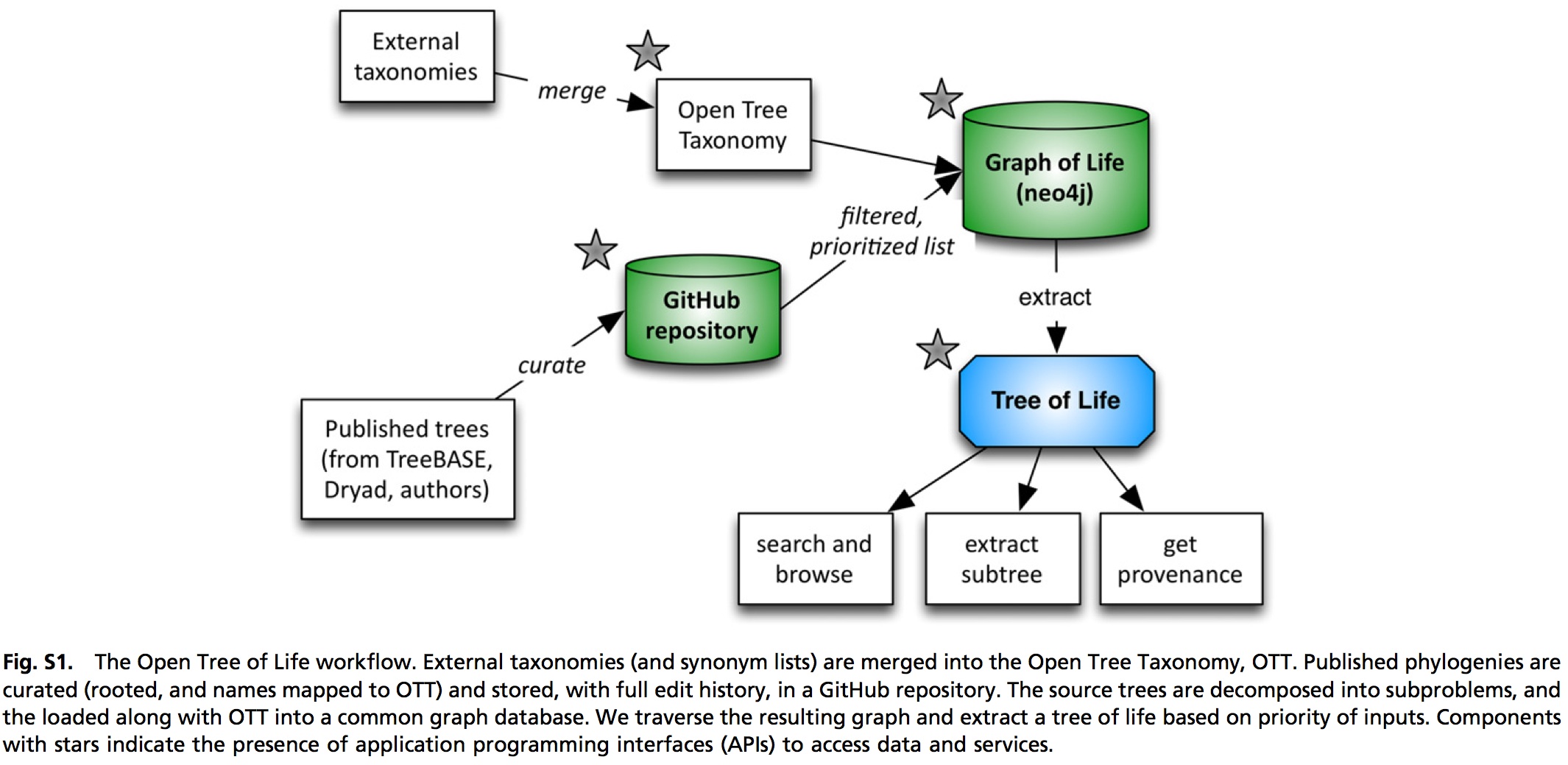
Tools to operate on large phylogenies
Given the increasing availability of large phylogenies for different taxonomic groups there is probably a tree out there that is fairly close to the set of taxa you’re interested in. Nonetheless, there are probably some additional steps to take when re-using such a tree:
- Taxonomic name resolution - for example, to reconcile the taxon names in the tree with those used in other data sets you already have (such as occurrence data, trait data, etc.).
- Pruning - to reduce the large input tree down to the set of taxa of interest, many superfluous taxa may need to be pruned out of the tree.
- Grafting - if a small number of taxa of interest is missing from the tree, there are algorithms that can place those taxa in the tree (at least, “close enough”) on the basis of the location of related species.

Taxonomic Name Resolution
When integrating data sets, you will often end up trying to reconcile taxonomic names from different data sources. Therefore, numerous data resources have APIs that allow for (fuzzy?) lookups of names, synonyms, alternative spellings:
- Encylopedia of Life
- Taxonomic Name Resolution Service
- Integrated Taxonomic Information Service
- Global Names Resolver
- Global Names Index
- IUCN Red List
- Tropicos
- The Plant List
- Catalogue of Life
- National Center for Biotechnology Information
- CANADENSYS Vascan name search API
- International Plant Names Index (IPNI)
- Barcode of Life Data Systems (BOLD)
- National Biodiversity Network (UK)
- Index Fungorum
- EU BON
- Index of Names (ION)
- Open Tree of Life (TOL)
- World Register of Marine Species (WoRMS)
- NatureServe
- Wikipedia
The taxize package allows you to scan all these different databases for name variants, common names, and higher classifications.
In this exercise, compare the outputs of NCBI and ITIS
Pruning and grafting
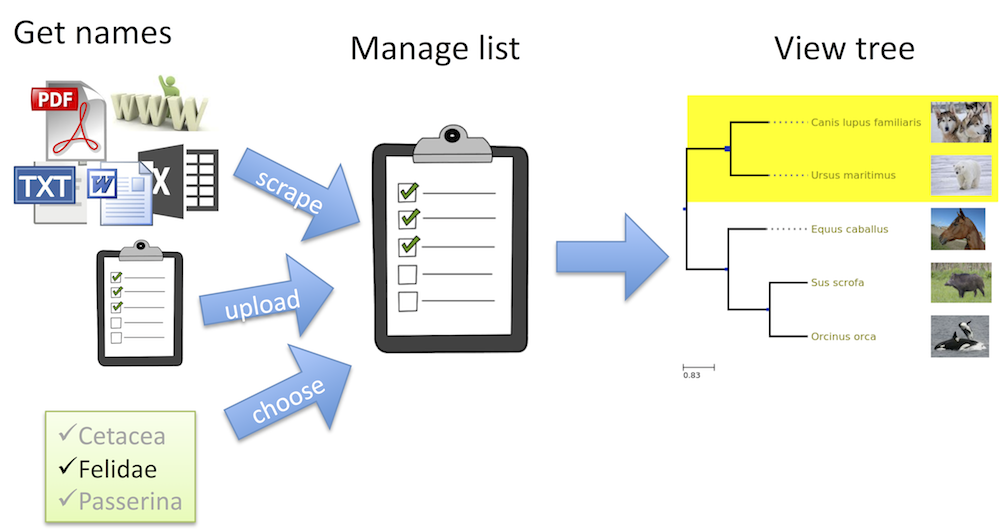
- Phylomatic was the first standalone tool and web
service that prunes large phylogenies down to the subtree for the provided input taxa, and
graft missing taxa by way of a path statement (e.g.
/ <family> / <genus> / <species>) that identifies where the taxon should be placed in relation to others. - Phylotastic is a re-design and re-implementation of the Phylomatic concept, but then with interchangeable components (e.g. tree stores, taxon services, pruning and grafting algorithms).
- S.PhyloMaker promises to infer trees and prune and graft supplied names, but it appears to be a bit buggy. Nevertheless, it does provide the very large PhytoPhylo reference tree.
In this exercise, prune the PhytoPhylo tree down to just our set of crop species, and inspect the subtree. What are some of the higher groups you recognize?