mebioda
Semantics
More especially: lexical semantics:
Examples of ambiguity in trait databases
When is the plant in bloom?
- BIEN:
plant flowering begin - TR8:
age_first_flowering - LEDA:
age_first_flowering - USDA:
Bloom_Period
How much do the seeds weigh?
- BIEN:
seed mass - TR8:
seed_mass,SeedMass,Seed.per.Pound - LEDA:
seed_mass - USDA
Seeds_per_Pound
The databases appear to have the same information - or close enough so that one type of
record (Seed.per.Pound) can be converted to another (perhaps
seed mass = 1lb. / Seed.per.pound). However, there are at least two challenges:
- Do we mean exactly the same? How do you define “flowering”?
- Are the units the same? If one observation is in “imperial” (pounds and ounces) and the other is in “metric” we have to know this if we are going to merge data.
We have an ontological problem
In computer science and information science, an ontology is a formal naming and definition of the types, properties, and interrelationships of the entities that really exist in a particular domain of discourse. Thus, it is basically a taxonomy.
An ontology compartmentalizes the variables needed for some set of computations and establishes the relationships between them.
The fields of artificial intelligence, the Semantic Web, systems engineering, software engineering, biomedical informatics, library science, enterprise bookmarking, and information architecture all create ontologies to limit complexity and organize information. The ontology can then be applied to problem solving.
What are some types of ontologies?
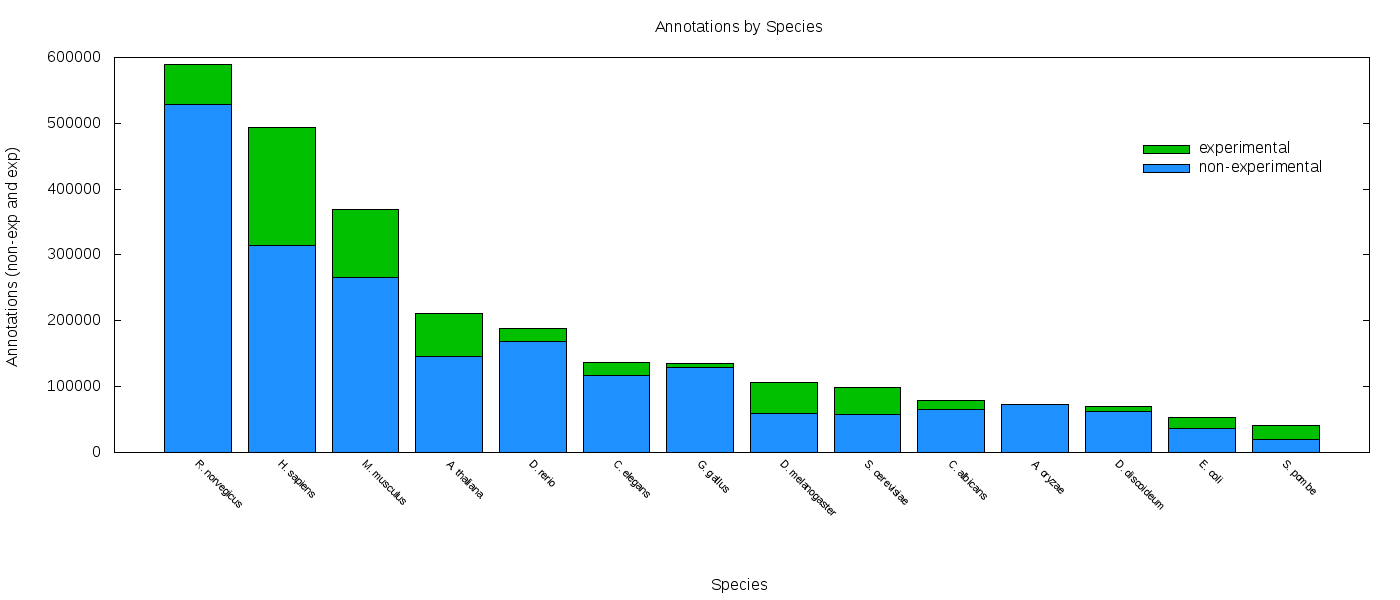
Data-driven ontologies have very many classes, which are (semi-)automatically generated from activities such as expression analyses (GO) or text-mining (FLOPO):
- The gene ontology
- The flora ontology
These types of ontologies can be used to do analyses such as enrichment tests, which assess whether a data set (for example, a list of genes that was expressed in a transcriptome) is significantly biased towards part of the ontology, such as a gene functional group (such as immune response).
Vocabularies to describe the essential concepts within a domain, such as:
Terms from such vocabularies are used to structure and annotate data. For example, the tabular occurrence records from GBIF use Darwin Core terms as column names. GenBank records use SO terms to label features on the sequence (such as exons, introns, CDSs).
Semantically annotated trait data
Data from the EoL trait bank is annotated with terms from the Darwin Core. We can query the database as follows:
library(traits)
library(taxize)
library(dplyr)
# example...
species <- "Cocos nucifera"
# For EoL traitbank data we need to provide the EoL taxon ID as input parameter. Hence,
# we first need to do a TNRS lookup of these, as follows:
sources <- gnr_datasources() # frame with global names sources
eol_id <- sources[sources$title == "Encyclopedia of Life", "id"] # lookup the id of the EOL source
eol_tnrs <- gnr_resolve(species, data_source_ids = c(eol_id), fields = "all") # resolve species
eol_taxon_id <- eol_tnrs[eol_tnrs$matched_name == species,]$local_id # lookup integer id
# Now that we have the taxon id, we query the traitbank
eol_results <- traitbank(eol_taxon_id)
eol_graph <- eol_results[["graph"]] # the interesting bit in the results is the graph
# Here we select the fields with Darwin Core terms
eol_triples <- select( eol_graph, 'dwc:scientificname', 'dwc:measurementtype', 'dwc:measurementvalue', 'units' )
# Write to file
write.csv(eol_triples, file = "eol_triples.csv")
Which produces this csv file
Any fact can be expressed in three parts
“A coconut will not
resprout following top (above ground
biomass) removal”
The three links can be viewed as the semantic anchors of this fact:
- the subject is coconuts: http://eol.org/1091712
- the predicate (think of this as the trait) is the ability to resprout: http://eol.org/schema/terms/ResproutAbility
- the object (in this case, the trait value) is the negation, the inability to resprout: http://eol.org/schema/terms/ResproutNo
Together, they form a triple:
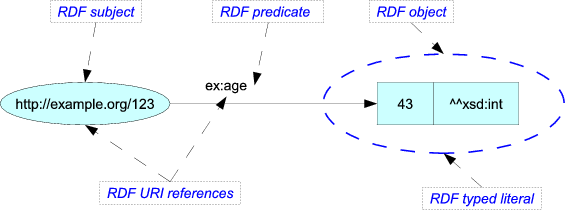
RDF
Our statement about coconuts, and all the other rows in our csv file can be reformatted in this simple RDF representation:
<http://eol.org/1091712> <http://eol.org/schema/terms/ResproutAbility> <http://eol.org/schema/terms/ResproutNo> .
By doing so, we integrate our data set in a web of knowledge representations, linking us to, for example:
- the plant trait ontology concept of ‘drought tolerance’
- the concept of ‘life span’ from the phenotypic quality ontology
- NASA’s understanding of human agriculture
All these triples together form a graph that, when published, can participate in the linked data graph:
RDF representation formats
RDF is fundamentally about abstract concepts (the triples thing), which can be
represented in many ways. The example above (<> <> <> .) is a terse form called
turtle, others include:
Large RDF data sets are typically not stored as files but in a special kind of database called a triple store (this is closer to the graph database neo4j used by OpenTree and EoL than to a relational SQL database).
Querying graphs: SPARQL and CYPHER
The language for querying relational databases (SQL) has a lot of functionality for expressing various types of joins and other clauses that combine tables. For graphs this is far less useful: emergent topological patterns are more important. Hence, languages for querying graphs have been developed. For RDF and the semantic web, the standard language SPARQL is endorsed by the w3c. For neo4j there is the (vendor-specific) language cypher.