mebioda
Big data phylogenetics
Outline
- Overview of the characteristics of phylogenetic data
- About representation formats
- Review of common data formats:
- Newick / New Hampshire
- New Hampshire eXtended (w. example analysis of TreeFam)
- PhyloXML
- Nexus
- NeXML (w. example analysis of TreeBASE)
- CDAO and RDF
- JSON
Phylogenetic data
- Plays a role in a variety of different contexts within biodiversity research, e.g.:
- Taxonomy and systematics
- Diversification analysis
- Comparative analysis
- Variety of different input data, e.g.:
- Multiple sequence alignments
- Morphological characters
- Computed distances (e.g. niche overlap?)
- Input trees (e.g. supertree methods)
- Variety of different methods, e.g.:
- Distance based, such as NJ
- Optimality criterion-based, such as MP and ML
- Bayesian, Markov chains
- Variety of different interpretations of output, e.g.:
- Tips are species, individuals, or sequences
- Branches are evolutionary change, time, or rates
- Nodes are speciations or duplications
- Topology represents evolutionary hypothesis or clustering
About representation formats
- Phylogenetic data is less ‘big’ than NGS data
- Often plain text-based, web standards (XML, JSON, RDF)
- More amenable to relational databases (SQL)
- Rarely represented in binary format (but see the examl parser for an application specific example)
The Newick / New Hampshire format
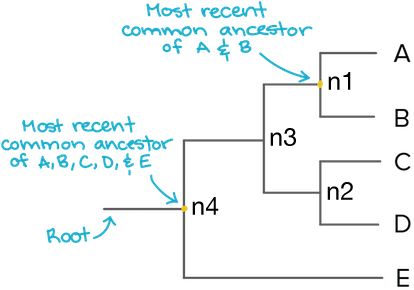
Newick representation:
(((A,B),(C,D)),E)Root;
- Optionally has branch lengths after each tip or node, as
:0.002321 - Optionally has comments inside square brackets, e.g.
[comment] - ‘Invented’ on a napkin at Newick’s Lobster House in Durham, New Hampshire, in 1986.
- Concise, but lacking all metadata
New Hampshire eXtended
- Format developed primarily for gene trees
- Additional data is embedded inside square brackets (i.e. should be backward compatible
with Newick format), which start with
&&NHX, followed by:key=valuepairs - Keys allowed:
GN- a text string, used for gene namesAC- a text name, for sequence accession numbersB- a decimal number, for branch support values (e.g. bootstrap)T- taxon identifier, a numberS- species name, a text stringD- flag to indicate whether node is a gene duplication (T), a speciation (F), or unknown (?)
Example:
(
(
( A[&&NHX:S=Homo sapiens], B[&&NHX:S=Homo sapiens] )[&&NHX:D=T],
( C[&&NHX:S=Pan paniscus], D[&&NHX:S=Pan troglodytes] )[&&NHX:D=F],
) , E
);
The technique to ‘overload’ comments in square brackets to embed data is also used in other contexts, such as:
- To store summary statistics of Bayesian analyses, as done by treeannotator
- To store branch and node decorations (e.g. color, line thickness), as done by Mesquite
Example of gene tree research: TreeFam data mining
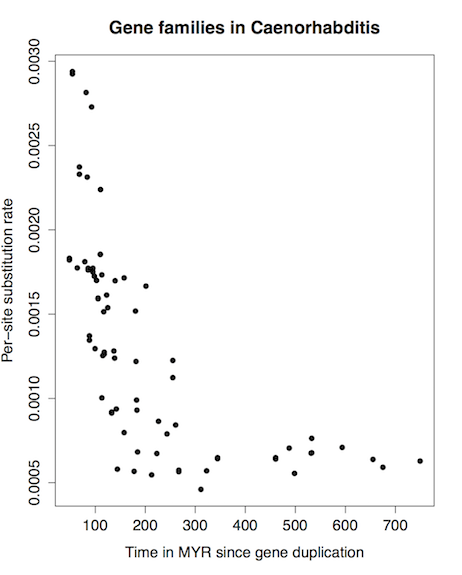
- Download the TreeFam data dump
- Extract and clean up NHX trees and FASTA data
- Perform fossil calibration on NHX trees (using a template command file for r8s)
- Extract rate as function of distance from duplication
- Draw a plot
PhyloXML
- Successor format to NHX
- Deals with the same concepts as NHX but in XML
<?xml version="1.0" encoding="UTF-8"?>
<phyloxml xmlns="http://www.phyloxml.org" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.phyloxml.org http://www.phyloxml.org/1.10/phyloxml.xsd">
<phylogeny rooted="true">
<clade>
<clade>
<clade>
<events><duplications>1</duplications></events>
<clade>
<name>A</name>
<taxonomy><scientific_name>Homo sapiens</scientific_name></taxonomy>
</clade>
<clade>
<name>B</name>
<taxonomy><scientific_name>Homo sapiens</scientific_name></taxonomy>
</clade>
</clade>
<clade>
<events><speciations>1</speciations></events>
<clade>
<name>C</name>
<taxonomy><scientific_name>Pan paniscus</scientific_name></taxonomy>
</clade>
<clade>
<name>D</name>
<taxonomy><scientific_name>Pan troglodytes</scientific_name></taxonomy>
</clade>
</clade>
</clade>
<clade>
<name>E</name>
</clade>
</clade>
</phylogeny>
</phyloxml>
The Nexus format

Nexus representation:
#NEXUS
begin taxa;
dimensions ntax=5;
taxlabels
A
B
C
D
E
;
end;
begin trees;
translate
1 A,
2 B,
3 C,
4 D,
5 E;
tree t1 = (((1,2),(3,4)),5);
end;
- Uses an extensible block structure that can also include character data (and other things, such as command blocks)
- Many different, mutually incompatible dialects that deviate from the original standard
- More facilities for metadata (e.g. names of things, annotations for taxa)
NeXML
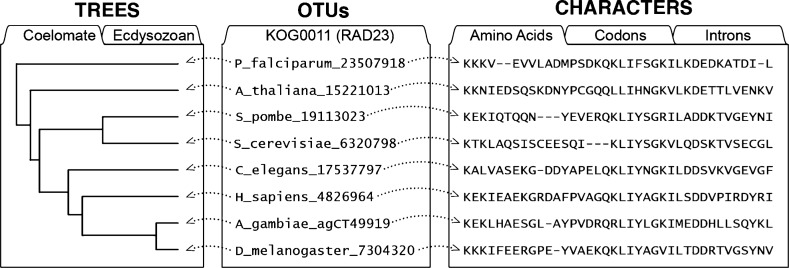
- Representation of Nexus as XML (example)
- The format in which TreeBASE’s data dump is made available
- Easily translatable, e.g. in R, python, perl:
#!/usr/bin/perl
use Bio::Phylo::IO qw'parse';
print parse(
-format => 'nexus',
-file => 'tree.nex',
-as_project => 1,
)->to_xml;
Example of species tree research: TreeBASE data mining

- Fetch the treebase sitemap.xml
- Download studies as nexml through the treebase API
- Convert trees to MRP matrices (Baum, 1992, Ragan, 1992) using a script
- Extract all species, and normalize the taxa
- Partition the data by taxonomic class
- Perform MP analyses with PAUP* and visualize the result (example: Arachnida)
This workflow was scripted using make for parallelization.
RDF and CDAO

- Facts can be represented as RDF triples
- Subjects and predicates are anchored on ontologies
- An example of this is the CDAO
- TreeBASE NeXML also has RDF statements in it, such as taxon IDs
- RDF can be queried with SPARQL
Tabular representations
- Unlike sequence data, trees are hierarchical data structures, which complicates tabular representation
- A simple table with child ID and parent ID columns works fine for small-ish trees, such as the Arachnida.csv that was the back end for the TreeBASE tree
- However, more general cases (such as networks) will require a node and an edge table
- Traversals require recursive queries, although some common queries (e.g. node descendants, ancestors, MRCA) can be implemented with additional, pre-computed indexes
// load the external data
d3.csv("Arachnida.csv", function(error, data) {
// create a name: node map
var dataMap = data.reduce(function(map, node) {
map[node.name] = node;
return map;
}, {});
// populate the tree structure
var root;
data.forEach(function(node) {
var parent = dataMap[node.parent];
if ( parent ) {
( parent.children || ( parent.children = [] ) ).push(node);
}
else {
root = node;
}
});
});